 |  |  |
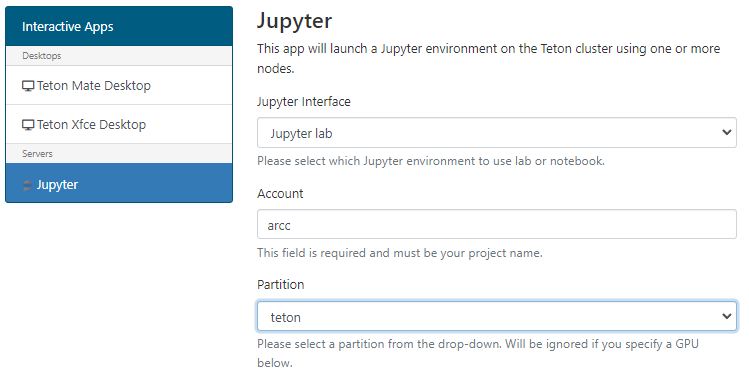
Define account / walltime and other required core/memory and GPU resources.
The maximum walltime allowed is 168 hours (seven days).
We would ask you to remember our general fairshare policy for cluster use and to try to request only the time you need and not to leave long sessions unused.
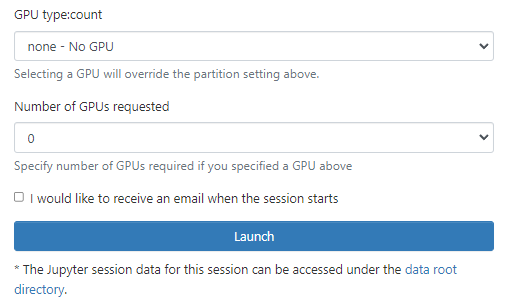
Wait for job to be allocated. You will see a message detailing that your job currently is sitting in the queue.
You will be allocated a node on the general cluster
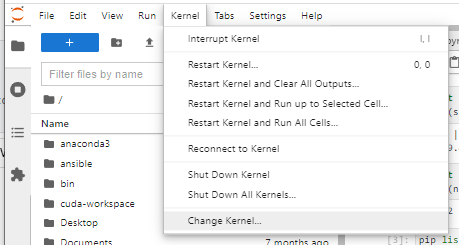
Click on the ‘Connect to Jupyter’ button to open a new browser tab to start your session.
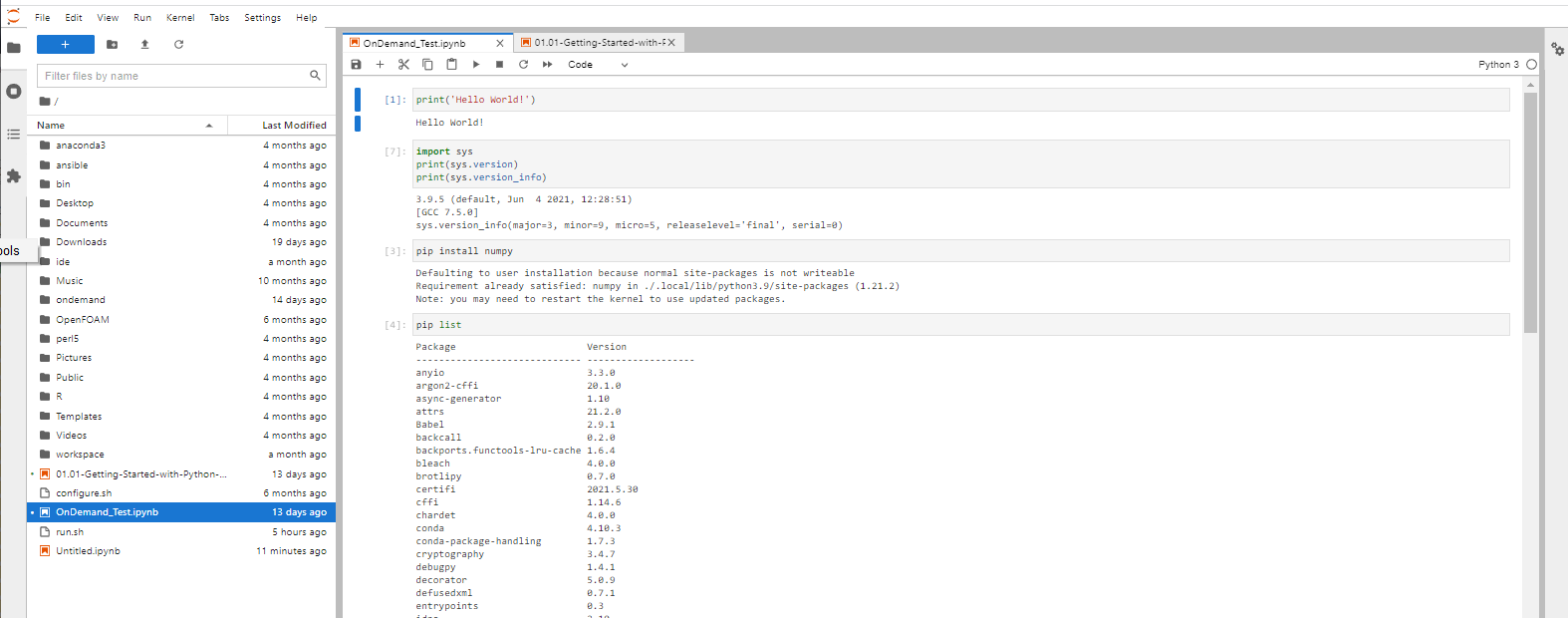
Use the notebooks.
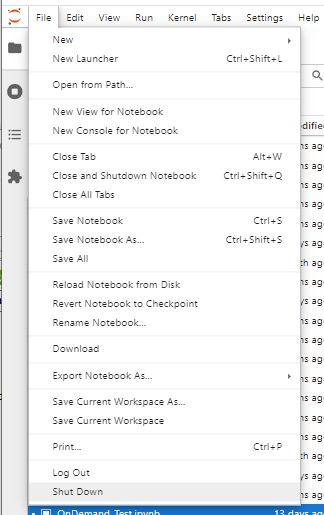
Once finished, shutdown your lab/notebook - this will cancel the underlying Slurm job, and free up resources.
Navigate to “My Interactive Sessions' to view the associated card.
You can delete this card or click on the Session ID to open a tab in the associated folder that was created in your home.
Your session will be allocated to an appropriate node on the cluster. If you’re logged onto the cluster you can perform squeue -u <username> to see your slurm allocation and which node it is running on.
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON) 2054747 beartooth sys/dash salexan5 R 1:32 1 t452 |
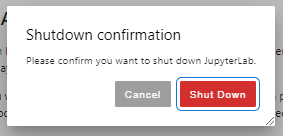
Once shutdown, your slurm allocation will be cancelled.
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON) |
Work related to this session can be found in the session’s related session id folder under the ondemand folder in your home folder. For example:
[salexan5@tlog2 output]$ cd /home/salexan5/ondemand/data/sys/dashboard/batch_connect/sys/jupyter/output [salexan5@tlog2 output]$ ls 13aea6e4-1966-4ee9-a86a-7b5ed55f6638 2d7afe4b-5f6f-44d5-b828-866ea4619fd6 5da92139-e166-42aa-8e46-03d851029940 839185fb-930a-4b16-b38d-37240002f42d f7c5b163-b689-417e-86d7-48e86d1b490e 1d74e3a5-332e-4588-a866-0e88d487b281 3bbda97d-7f59-4ebd-9b7b-91b5334ab560 61637eb8-db59-4af3-8f7a-e48ab33ee6ca 8d9d1a5f-89a4-4386-a89a-74854d9ba346 faec06c1-7e88-40c3-a4c8-af49c520b5eb 1fd86d70-6187-4a28-9dc0-78bcd998c2e9 3d3905a6-500e-4227-8658-b3a1ffc4d0b6 6b23acf2-1a14-4bac-9f2a-266b55a577d0 d42426a6-308d-48f1-ab71-bad01012609e 2127f26c-dfae-4905-a984-0f5753655309 48f98884-e59b-495b-b1a8-800c5c996ede 79fc87b2-c96e-4ff8-bc30-b393fd897bda f471ae5f-2b28-4858-bf9c-925d4209e277 [salexan5@tlog2 f471ae5f-2b28-4858-bf9c-925d4209e277]$ ls after.sh before.sh config.py connection.yml job_script_content.sh job_script_options.json output.log script.sh user_defined_context.json |
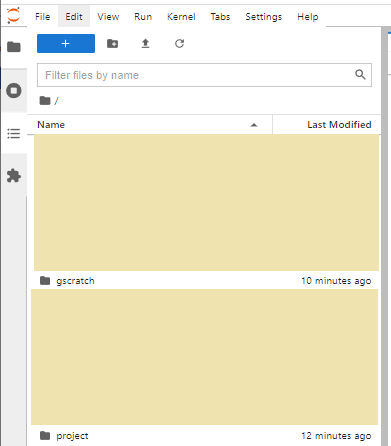
Currently you can not directly navigate out from you /home into your /project nor /gscratch.
The suggested solution to this is to create symbolic links to you project and gscratch, within your home, that you can then click on to navigate.
For example:
[~] ln -s /project/arcc project [~] ln -s /gscratch/salexan5/ gscratch [~] ls -al ... lrwxrwxrwx 1 salexan5 salexan5 19 Nov 4 16:15 gscratch -> /gscratch/salexan5/ ... lrwxrwxrwx 1 salexan5 salexan5 13 Nov 4 16:13 project -> /project/arcc ... |
When you start a new Jupyter session, these two symbolic links will be your file manager. Double clicking on these folders will navigate appropriately:
The answer is yes, and what follows is based on NCAR CISL’s page on Using conda environments for Python access.
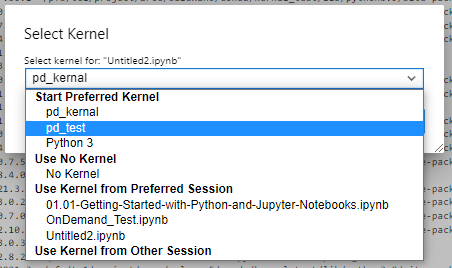
If you’re creating a new, or have an existing conda environment you can add the ipykernel package, and then configure the environment to be consider as a kernel.
The following example creates a python 3.10 environment with Pandas, within my project folder, which I want to be able to use in Jupter.
# navigae to my project space. [salexan5@tlog2 ~]$ cd /project/arcc/salexan5/conda/ [salexan5@tlog2 conda]$ module load miniconda3/4.10.3 # Use the -p option to create the conda environment in my current folder, rather than my home. [salexan5@tlog2 conda]$ conda create -p pandas_test python=3.10 ... # To activate this environment, use # $ conda activate /pfs/tc1/project/arcc/salexan5/conda/pandas_test # To deactivate an active environment, use # $ conda deactivate ... [salexan5@tlog2 conda]$ conda activate /pfs/tc1/project/arcc/salexan5/conda/pandas_test # This step is used to force conda to install all packages specifically within the environment, and not within any local or other locations. (/pfs/tc1/project/arcc/salexan5/conda/pandas_test) [salexan5@tlog2 conda]$ export PYTHONUSERBASE=intentionally-disabled (/pfs/tc1/project/arcc/salexan5/conda/pandas_test) [salexan5@tlog2 conda]$ conda install -c conda-forge pandas ... (/pfs/tc1/project/arcc/salexan5/conda/pandas_test) [salexan5@tlog2 conda]$ conda install ipykernel ... (/pfs/tc1/project/arcc/salexan5/conda/pandas_test) [salexan5@tlog2 conda]$ python -m ipykernel install --user --name=pd_test Installed kernelspec pd_test in /pfs/tc1/home/salexan5/.local/share/jupyter/kernels/pd_test # Note the location in your home folder where this configuration is stored. # Within this folder you'll find the kernel.json file that points to the location of this conda environment. (/pfs/tc1/project/arcc/salexan5/conda/pandas_test) [salexan5@tlog2 conda]$ python --version Python 3.10.2 (/pfs/tc1/project/arcc/salexan5/conda/pandas_test) [salexan5@tlog2 conda]$ conda list # packages in environment at /pfs/tc1/project/arcc/salexan5/conda/pandas_test: ... ipykernel 6.7.0 pypi_0 pypi ... numpy 1.22.1 pypi_0 pypi ... pandas 1.4.0 pypi_0 pypi ... (/pfs/tc1/project/arcc/salexan5/conda/pandas_test) [salexan5@tlog2 conda]$ conda deactivate |
Notice the name of kernel is pd_test, the version of python, and the versions of numpy (1.22.1) and pandas (1.4.0).
Start a Jupyter session and then: